Three-Dimensional Point Tracking¶
These scripts allow for the easy tracking of blobs embedded in vortex paths. They rely on a number of modules (some internal, some developed out-of-house) and assume a certain level of processing has occurred prior to their use. This documentation will walk you through how to download and install the modules you’ll need, explain how the code works, and walk you through some example usage using test data included with the scripts.
Contents and Dependencies¶
Tracking scripts are accessible via the git server. This folder also includes sample data to test each of the scripts. To download, run the following command (this will require a log-in to the Git server):
$ git clone ssh://<YOUR_NAME>@immenseirvine.uchicago.edu/git/tracking
The cloned folder should contain three scripts that will be necessary for tracking the dye blobs:
- process_encoder_speeds.py
- track_dye_blobs.py
- process_blob_data.py
The first script extracts cart velocity data from the encoder output (note that this is also required for the tracing of the paths, so it will often be the case that this step is already complete when you’re ready to track the blobs – more on this later). The second script is responsible for tracking the blobs in space and recording particular features of their locations at a given frame, while the final script computes relevant quantities from that information. There should also be another python script, sparse.py, in the folder which will be imported by the other python scripts. This code holds the class for handling sparse4d objects.
To use these scripts, you will need two modules in ILPM that make handling vectors and paths simple and straight forward:
- vector
- path
You will also need the following modules developed by external sources:
- pandas
- trackpy
trackpy is responsible for actually tracking the blobs once they have bene identified, while pandas provides a spreadsheet-like object in which the data is stored. See the following section for details on acquiring and installing these modules.
Module Installation¶
ilpm package
To download the ILPM package, simply check it out from the git repo and use the setup.py file to install it:
$ git clone ssh://<YOUR_NAME>@immenseirvine.uchicago.edu/git/ilpm
$ cd ilpm
$ sudo python setup.py develop
After doing so, you should have global access to the modules contained in the package. You can check your access to vector and path modules by navigating to a directory that does not contain the ILPM folder, entering a python environment and typing:
from ilpm import vector, path
If no error is returned, the installation was successful and you now have access to all ILPM modules using the same import syntax.
pandas
This module is well documented and well maintained by the python community. As such, it’s no surprise that it can be easily pip installed. To download and install the module globally, simply type:
$ sudo pip install pandas
trackpy
This module is maintained by an outside group of scientists. There are a number of ways to download and install this module (listed here: http://soft-matter.github.io/trackpy/v0.3.2/installation.html ), however, pip is an easy way of doing it without requiring you to download additional packages. To download and install trackpy, use:
$ git clone https://github.com/soft-matter/trackpy
$ pip install -e trackpy
Note that the trackpy documentation also suggests you download and install a number of other modules – with the exception of pandas, these are not necessary to use the scripts included in the tracking git folder, but may be necessary for the use of the full trackpy module.
Required Directories and Files¶
The two scripts included in the tracking directory require certain files to exist before they’re able to successfully track the blobs and report relevant information. Here is information on what these scripts are expecting (note that the test_data directory contains exactly the minimal starting information, so that should serve as a nice reference as well):
Directory Structure
The basic data input for these scripts is the .sparse file that corresponds to a particular experiment. This sparse file is expected to exist in the same directory as the corresponding .s4d file. Additionally, there should be a folder at the same level of the .sparse and .s4d files that is named by the date in YYYY_MM_DD format, which contains information regarding the cart kinematics. Finally, there should be a directory with the same name as the sparse file, which will serve as the main directory for all the processing results.
Thus, the minimal directory structure should look like this:
2017_02_01
2017_02_01_my_first_experiment
2017_02_01_my_first_experiment.sparse
2017_02_01_my_first_experiment.s4d
Required Pre-processing
In addition to assuming some basic high level directory structure (which was likely automatically created by the tracing scripts), there are a few processes that need to be performed for the scripts to successfully track and interpret the information about the blobs. Here’s a run down of the required information:
Vortex Paths: The vortices need to be traced before the blobs can be tracked. The resulting paths should be stored as .json files in a subdirectory called paths in the directory named after the sparse file, e.g.: 2017_02_01_my_first_experiment/paths/0000.json.
Circulation Calibration: The circulation of each vortex is inferred based on a circulation calibration curve. These curves are generated by an independent set of PIV measurements, that measure the circulation of the vortex against the final speed of the cart. Fitting the resulting data to a line produces two parameters: the slope of the fit line and its intercept. To process the blob information with process_blob_data.py, you will need to pass each of these numbers to the code as a command line argument.
Cart Speed: To get the circulation of the vortex, the code will need to know the final cart speed. It will look for this information in the directory named after the date and will assume that the data has the form returned by process_encoder_speeds.py, which needs to be run on that directory prior to processing the blob tracks. Example usage would be:
$ python process_encoder_speeds.py /Volumes/labshared2/<USER>/2017_02_01_experiments/2017_02_01/
Tracking Process, Step-by-step¶
Here we break down the code contained in track_dye_blobs.py to illustrate the functionality of each chunk of code. Note that the code itself also contains inline comments intended to guide the reader through the processing.
Organize Files and Directories
There are a number of outputs generated during the tracking process, some of which are diagnostic and others are the desired data. These files need to be named correctly and stored in the appropriate directories, so the first thing the code does is ensure the directory structure is in place.
#ENSURE PROPER DIRECTORY STRUCTURE EXISTS
base, sparse_name = os.path.split(sparse_file)
name = os.path.splitext(sparse_name)[0]
s4d_name = name+'.s4d'
s4d_file = os.path.join(base,s4d_name)
S4D = cine.Sparse4D(s4d_file)
setup_info = S4D.header['3dsetup']
blob_dir = os.path.join(base,name,'blob_proj')
if not os.path.exists(blob_dir): os.mkdir(blob_dir)
peak_dir = os.path.join(base,name,'blob_peaks')
if not os.path.exists(peak_dir): os.mkdir(peak_dir)
path_dir = os.path.join(base,name,'paths')
if not os.path.exists(path_dir):
print "!! Paths need to be traced in order to track blob motion !!"
continue
tracking_dir = os.path.join(base,name,'blob_tracking')
if not os.path.exists(tracking_dir): os.mkdir(tracking_dir)
Sort Traced Paths
Following the tracing procedure, there is no guarantee that the first path in the tangle is always the same physical path, or that the same number of paths will be traced for each volume. To get around this, the code will go through all the paths and use some specified criteria to assign consistent path numbers to any paths present in a volume. Currently, length and writhe are supported criteria for assigning path numbers; however, you could edit the code to implement any criteria you felt was robust. The output of this, frames_numbered_paths, is a list of arrays containing the paths, where the index represents the path number and a None will appear wherever that path disappears for a frame.
#FIND AND SORT ALL TRACED PATHS
sorted_frames, sorted_json_files = get_sorted_frames_jsons(path_dir)
frames_numbered_paths, path_number = make_path_number_assignments(path_dir, sorted_frames, sorted_json_files, methods=['length'])
Isolate Vortex Volume
The first step in the tracking process is to use a slightly smoothed version of the the traced paths smoothed_X to isolate a sub-volume subVol centered on the vortex from the s4d volume V. An arbitrary framing for the path is generated and then used to create a mesh that is regular in that basis. The volume is then sampled on this mesh, returning a rectilinear array whose axes correspond to the framing basis. The first axis (tangent vector) will have a length set by how densely the path is resampled (default NP= 1000). The extent of the transverse directions, i.e. the second and third axes (normal, binormal vector), is set by the global variable AREA, which measures the half length of the volume’s cross-section in voxels.
#LOAD THE VOLUME
V = S4D[f].T
#SMOOTH THE PATH AND SELECT THE SUBVOLUME AROUND THAT PATH
smoothed_X = smooth_path(X, path_smoothing)
subVol, Xi, s = extrude_path_centered_volume(smoothed_X, V, setup_info, area=AREA)
Identify Intensity Maxima
Once the volume around the vortex has been isolated, it is collapsed in the transverse directions by summing over those axes, producing collapsed_intensity. The resulting 1D intensity profile is then smoothed using periodic boundary conditions and then the position of the local maxima are determined. smoothed_intensity is then used to get the (x,y,z) coordinates of the blobs in space blob_locations. The information about each blob for a single frame is stored as an entry in a dictionary called blob_info.
#COLLAPSE SUBVOLUME AND IDENTIFY LOCAL MAX'S IN THE INTENSITY PROFILE
collapsed_intensity = subVol.sum(-1).sum(-1)
smoothed_intensity = ndimage.gaussian_filter1d(collapsed_intensity, SIGMA, mode='wrap')
peak_indicies = locate_peaks(smoothed_intensity)
#USE INTENSITY PEAK LOCATIONS TO GET BLOB INFORMATION
blob_locations, blob_path_positions, blob_tangents = back_out_blob_info(peak_indicies, Xi, s, setup_info)
#STORE BLOB INFORMATION FOR THIS FRAME-PATH COMBINATION
blob_info[f] = {'blob_locations': blob_locations,
'blob_path_positions': blob_path_positions,
'blob_tangents': blob_tangents,
'L': sum(vector.mag(vector.plus(X)-X)),
'H_c': sum(path.Tangle([X]).crossing_matrix()[0]),
'X': X
}
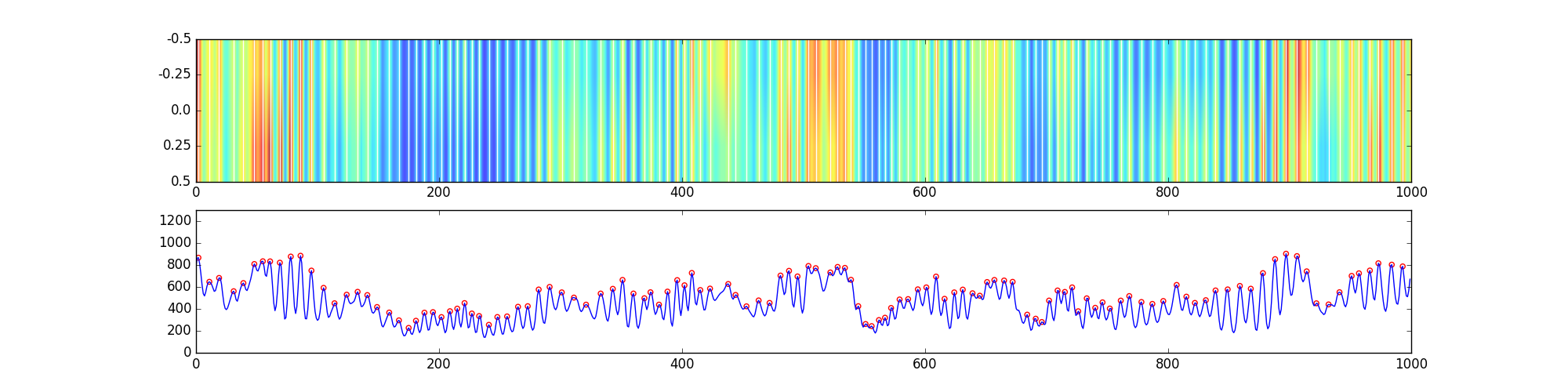
A summary of this processes is stored as a figure in the blob_proj subdirectory, an example of which is shown below. The top panel shows the subVol intensity summed along a single direction, with peak locations overlaid as white lines. The bottom panel shows the subVol data summed over both transverse axis and smoothed, with peak locations indicated by open red circles.
Track the Blob Positions
After the blobs in each volume have been identified, the entirety of the blob information is passed to trackpy, which then links blobs across frames to produce tracks. There are two relevant parameters in this process:
- max_disp: When trying to establish a correspondence between blobs in adjacent frames, trackpy will only consider blobs within a given distance of the current blob when searching for partners. max_disp sets the distance over which trackpy will search. It’s best to have this number be as small as possible and still support the dynamics of the system, since a larger number both increases the computation time and the likelihood that mistakes in tracking are made.
- memory: trackpy is tolerant to blobs blinking in and out of the movie. If a blob vanishes for a frame and reappears in a later frame, trackpy is able to recognize the two images as the same blob. memory sets the maximum number of frames a blob is allowed to disappear for and still be considered the continuation of a previously established track.
The result of the tracking is a data frame pandas object that contains the positions and ID’s of each of the blobs, along with additional information about each blob at a particular frame, including the tangent at that point.
#TRACK THE BLOBS ON THIS PATH, SAVE THE RESULTS AS A DATAFRAME
tracking_df_fn = os.path.join(tracking_dir, 'path_%d.pickle'%i)
if os.path.exists(tracking_df_fn) and not overwrite_tracks: continue
else: track_blobs_from_peaks(blob_info, tracking_df_fn, max_disp, memory)
Running Scripts on Test Data¶
On immense in labshared2, there is a directory called test_data, which has a subdirectory called blob_tracking. This folder contains all the sample data needed to run the scripts contained in tracking. Inside the directory, you should find the following:
$ 2015_05_12
$ 2015_05_12_THL001_psi40_spacer7_filter_over_solohelix_eye_II
$ 2015_05_12_THL001_psi40_spacer7_filter_over_solohelix_eye_II.sprase
$ 2015_05_12_THL001_psi40_spacer7_filter_over_solohelix_eye_II.s4d
The first directory contains the output from the optical encoder, while the second contains a subdirectory that contains the paths for this particular sparse file.
To start, you’ll need to parse the output from the encoder to make it readable by later steps in the tracking code. Note that generating the paths also requires that this encoder information be processed, and if that is the case for your data, you don’t need to do it again. Processing the data is accomplished using process_encoder_speeds.py:
$ python process_encoder_speeds /Volumes/labshared2/test_data/blob_tracking/2015_05_12/
Check that after running this script there are a number of .pickle files in addition to .txt and .png files in the encoder directory now. The next step is to identify and track the blobs. This is done with a single call:
$ python track_dye_blobs.py /Volumes/labshared2/test_data/blob_tracking/2015_05_12_THL001_psi40_spacer7_filter_over_solohelix_eye_II.sparse
Note that there are number of optional command line arguments for this call:
- -path_smoothing: The percent by which the path is smoothed before extracting a subvolume. The default is 1%.
- -max_disp: The maximum displacement allowed by a blob in tracking the blobs. The default is 5 voxels.
- -memory: The memory limit used during tracking. The default is 2 frames.
- –overwrite_peaks: If included, will identify peaks on frames where peak information already exists.
- –overwrite_tracks: If included, will track the particles and overwrite existing dataframes.
The process should produce a number of subfolders under the 2015_05_12_THL001_psi40_spacer7_filter_over_solohelix_eye_II directory, including:
- blob_proj
- blob_peaks
- blob_tracking
At the end of the process, there should be snapshots of the peak identifications in blob_proj, and a single file in each of the other two directories, which stores the information about the blobs over time for the single path in the sparse. A typical snapshot of the peaks is included above in the section discussing peak identification.
To process the tracks, a single call to the final python script is made:
$ python process_blob_data.py /Volumes/labshared2/test_data/blob_tracking/2015_05_12_THL001_psi40_spacer7_filter_over_solohelix_eye_II.sparse -birth_frame 9 -gamma_calibration 13.9 1527
This script calculates useful quantities from the tracked blobs. It requires two additional inputs: first, the birth_frame, which is the frame of the s4d during which the vortex is actually generated, and second, the gamma_calibration, which is the slope-intercept pair that defines the cart-speed vs. circulation calibration.
This script will create two subfolders under the 2015_05_12_THL001_psi40_spacer7_filter_over_solohelix_eye_II directory:
- local_measures
- geometric_measures
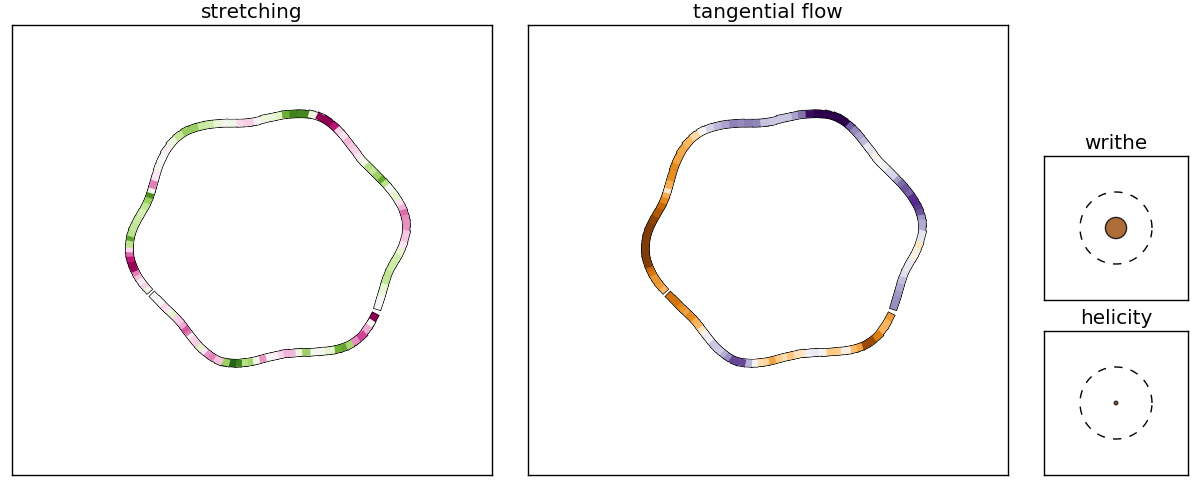
The first of these contains snapshots of the local stretching and tangential flow on the vortex, and also shows how the current total helicity compares to the current writhe (the dashed circle is a writhe or helicity of 1.0). A sample plot for frame 19 is shown below:
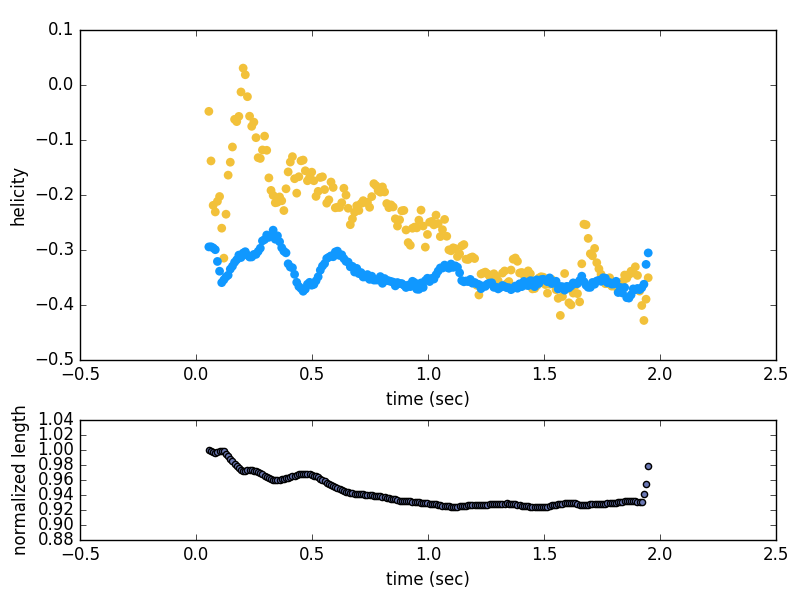
The geometric_measures folder contains the aggregated information for the writhe, helicity, and length over the evolution of each path. It will also make a figure showing how these quantities evolve over time. The output for the test data is shown below: